 Marine animals need to control osmotic pressure within their cells, especially in the context of their saline environment (the sea). Two organic biological molecules play a significant role in this: Urea and TMAO.
Marine animals need to control osmotic pressure within their cells, especially in the context of their saline environment (the sea). Two organic biological molecules play a significant role in this: Urea and TMAO.
Both TMAO and urea act as osmolytes, regulating osmotic pressure and ensuring cell structure and ion concentrations are maintained in the marine body. These organic molecules also interact with proteins but with opposing effects.
Urea is a denaturant, which means that it can alter the structure of proteins by weakening and breaking bonds. This can lead to the 3D structure of the protein being changed and its function being lost. Conversely, TMAO prevents the denaturing of proteins, and it has been suggested that TMAO interacts with urea, acting to reduce its denaturation effects.
The mechanism behind urea-induced denaturation is well understood, however there is still little knowledge on the mechanism behind the protective effects of TMAO on proteins. This study aimed to increase understanding of TMAO and its role in urea depletion at a protein surface, as well as reveal the atomic interactions between TMAO, urea and a tripeptide glycine–proline–glycinamide (GPG) for the first time. The research builds previous work at ISIS which used neutron diffraction to study TMAO interactions with water at both high (4 kbar) and ambient pressure (25 bar).
Neutron diffraction on NIMROD was used to study the atomic interactions between TMAO, urea and GPG, which is a model structural motif found in proteins. GPG was studied under three conditions: aqueous TMAO, aqueous urea and a 2:1 ratio of aqueous TMAO and urea.
Their results showed that a network of weak hydrogen bonds is formed between TMAO, urea and water. This results in urea being depleted from the surface of GPG. In the case of the TMAO and urea samples, this bonding was three times more likely to occur between TMAO and urea compared to TMAO and water, which further suggests that TMAO acts to reduce the denaturation effects of urea by preventing urea from moving towards the peptide surface.
This research clearly demonstrates that TMAO has unique properties regarding protein interactions as it counteracts urea-induced denaturation by incorporating urea into a hydrogen bond network in solution. Further research is needed to confirm this observation occurs in larger and more complex proteins.
The project was supported by a grant from the Engineering and Physical Sciences Research Council (EPSRC) (EP/ P02288X/1) and a European Research Council Consolidator Fellowship/ UKRI Frontier Research Fellowship for the MESONET project UKRI (EP/X023524/1). The paper can be found here: Trimethylamine-N-oxide depletes urea in a peptide solvation shell | PNAS
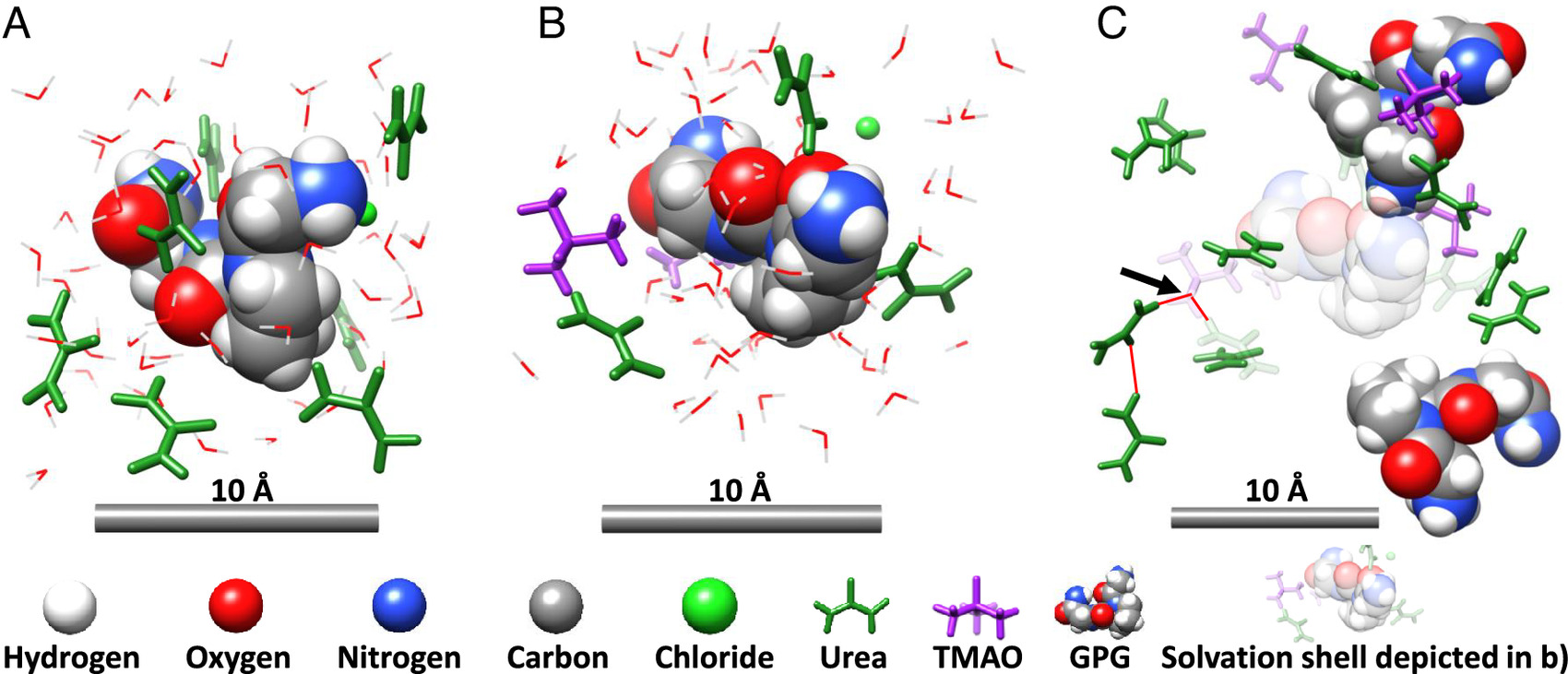
Representative solvation structures drawn from the structural refinement modeling of the diffraction data that pertain to the peptide in aqueous urea (A) and aqueous urea–TMAO (B and C). (A) shows the urea-rich surface of GPG tripeptide in aqueous urea. (B) shows the surface of a GPG tripeptide that is now relatively depleted in urea, in a solution of aqueous urea–TMAO, and (C) shows the bulk-solution further surrounding the GPG interface of the structure in (B) in aqueous urea–TMAO. The arrow marks a TMAO molecule entrapping urea in a hydrogen bond network. Urea and TMAO molecules are colored green and purple throughout, while water molecules are described by a wire form which has been hidden in (C) to aid visualization.
